Research
Monarch Initiative: Disease diagnosis and discovery

Over the last 20 years, the genomic revolution has provided tremendous amounts of knowledge in nearly all scientific fields, especially genomic medicine. But that data is complex, exists in different forms, and is distributed across different locations. It can also be incomplete or conflicting, or downright overwhelming to identify what parts of available knowledge are relevant to any given question.
The challenges right now are not due to lack of data–there is a huge and growing amount of data. What we lack are tools to make effective use of that data for everyone. The Monarch Initiative was recently expanded through a $10M award to establish a Center of Excellence in Genomic Sciences (CEGS) and develop tools to address exactly this data problem. Through the CEGS project, scientists and physicians at four institutions in the U.S. and U.K are collaborating to develop those tools.
Monarch is a collaboration among members TISLab, Lawrence Berkeley National Laboratory, The Jackson Laboratory For Genomic Medicine, RTI International, Genomics England/Queen Mary, Ada Health, and EMBL-EBI
Contact: Melissa Haendel
Funding: NIH grant # 1R24OD011883-01 and NHGRI 1RM1HG010860-01
Phenomics-First Resource
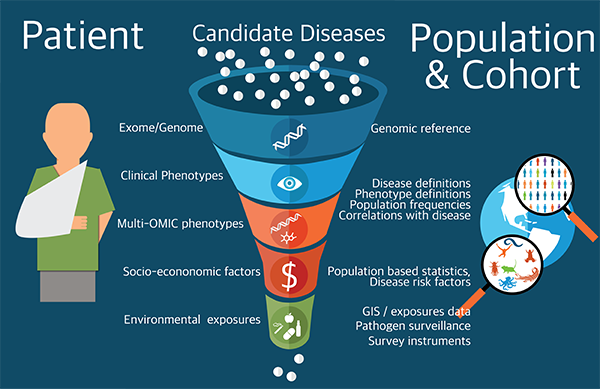
The human genome has been sequenced, and yet so much is unknown about what it does; what we do know is scattered across multiple, heterogeneous data sources that are difficult to integrate. The Phenomics-First Resource (PFR) coordinates the efforts of a large community of researchers and clinicians to help interpret the relationship between the differences (called variants) in a patient’s genome and their physical characteristics (phenotypes). We are using the emerging field of phenomics to improve precision medicine and innovate in evolutionary biology. Patients with rare diseases will particularly benefit from this research; it takes an average of seven years to get a diagnosis for a rare disease, and even then the initial diagnosis is often wrong. Phenomics can be applied in a range of fields, but in this case the researchers are primarily focused on human disease and drawing insights from laboratory organisms.
The PFR will gather and unify the data from numerous sources to advance genomic interpretation in clinical settings, and will empower both researchers and clinicians to overcome long-standing barriers to discovery and patient care. This project forms the basis of a Center of Excellence in Genomic Science, a program of the National Human Genome Research Institute (NHGRI), from the National Institutes of Health (NIH).
Contact: Melissa Haendel
Funding: NHGRI
Center for Linkage and Acquisition of Data
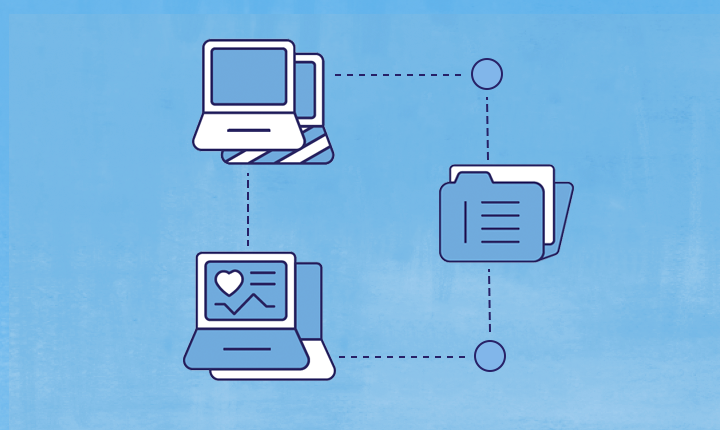
The All of Us Center for Linkage and Acquisition of Data (CLAD) expands the types and research utility of program data available to researchers through passive data streams. CLAD acquires and processes linked data within a secure platform before delivering it to the All of Us Data and Research Center. The CLAD team is also responsible for developing analytical tools to help researchers jumpstart their analyses when using the linked data.
Contact: Anita Walden
Funding: NIH Office of the Director, # OTA-23-003
INCLUDE Data Coordinating Center

The INCLUDE Data Coordinating Center (DCC) is creating a cloud-based, open, digital platform to easily access and share Down syndrome resources to maximize and accelerate research.
Contact: Melissa Haendel
Funding: Supported by the National Institutes of Health INCLUDE Project under Project Number U2CHL156291 administered by the National Heart, Lung and Blood Institute.
Biomedical Data Translator Program

NCATS studies translation on a system-wide level as a scientific and operational problem. TISLab convenes expert teams from diverse scientific disciplines — including, among others, efficacy, toxicity, data sharing, biomarkers and clinical application — to reduce, remove or bypass significant bottlenecks across the entire continuum of translation and to train the future translational science workforce.
Contact: Matt Brush
Funding: NCATS grant # 3 OT3 TR002019 01S2
National COVID Cohort Collaborative (N3C)

The National Center for Advancing Translational Sciences (NCATS) and the National Center for Data to Health (CD2H) have created the National COVID Cohort Collaborative (N3C) Data Enclave. The N3C aims to aggregate, harmonize, and make accessible vast amounts of data from healthcare providers nationwide to accelerate advances in COVID-19 research and clinical care. With the uncertainty of the COVID-19 global pandemic, the scientific community and Clinical and Translational Science Awards (CTSA) Program created the N3C partnership to overcome technical, regulatory, policy, and governance barriers to harmonizing and sharing individual-level clinical data.
N3C welcomes researchers and citizen scientists to sign up and register for access to this nationwide, comprehensive health records dataset. Learn more at the CD2H N3C page.
Contact: Melissa Haendel
Funding: National Institutes of Health’s National Center for Advancing Translational Sciences, Grant Number U24TR002306
NHLBI BioData Catalyst® (BDC)
The NHLBI BioData Catalyst® (BDC) is a cloud-based ecosystem that offers researchers data, analytic tools, applications, and workflows in secure workspaces. It is a community where researchers can find, access, share, store, and analyze heart, lung, blood, and sleep data resources. And it is one of NHLBI’s data repositories, where researchers share scientific data from NHLBI-funded research so they and others can reproduce findings and reuse data to advance science. By increasing access to NHLBI data and innovative analytic capabilities, BDC accelerates reproducible biomedical research to drive scientific advances that can help prevent, diagnose, and treat heart, lung, blood, and sleep disorders. TISLab, in collaboration with RTI International, is leading the Data Management Core (DMC) for BDC. The goal of DMC is to streamline the process of submitting, ingesting, and harmonizing data in BDC.
Contact: Anne Thessen
Funding: Funding is National Heart, Lung, and Blood Institute OT2HL167310
Advancing a Community-led Zebrafish Toxicology Phenotype Atlas (ZAPP)
Zebrafish have greatly advanced toxicology and environmental studies as a valuable animal model because they are easy to manipulate, breed, and observe during development. However, the absence of universal standards significantly impedes scientific progress and, therefore, human and environmental health.
In this project, we develop standard ontologies and data models to improve data compatibility within and across species and domains. We create a toolkit allowing community members to annotate their phenotypic data, mainly created through toxicological assays, to generate “born-interoperable” data.
Finally, we create a zebrafish phenotype atlas web application. This atlas serves as a visual definition of the standards and their documentation for examining variations of specific phenotypes by laboratories in the community. Users will be able to explore and query toxicological exposures and phenotypes of interest and see example images demonstrating the phenotypes.
This project is a collaboration with Alexa Burger’s team at the University of Colorado Anschutz Medical Campus, and community-governed by diverse stakeholders in toxicology and environmental health sciences.
Contact: Sabrina Toro
Funding: Funding is NIEHS - R24ES036130
LinkML: an open data modeling framework

LinkML (Linked data Modeling Language) is an open, extensible modeling framework that allows computers and people to work cooperatively to model, validate, and distribute data that is reusable and interoperable. It is designed to create interoperable data from the start without the overhead normally required for doing this. The LinkML user-requested features that will be addressed by this funding are:
- Support our growing open-source developer community (SUPPORT)
- Lower the bar to creating LinkML models (CREATE)
- Extend model transformations (EXTEND)
Contact: Matt Brush
Funding: Funding is Chan Zuckerberg Initiative CZI 313291/Z/24/Z
Past Research
Gabriella Miller Kids First Pediatric Data Resource Center

TISLab is fighting pediatric cancer by supporting The Kids First Data Resource Portal, one of the largest collections of integrated genomic and clinical data for some childhood diseases, which previously were studied largely in isolation. Kids First brings together heterogeneous data from childhood cancers and structural birth defects to support research, study, and collaboration built on top of an unprecedented collection of genetic and phenotypic data from pediatric patients. The data in the Data Resource Portal is curated and structured using biomedical ontologies, including the Human Phenotype Ontology (HPO) and Mondo Disease Ontology to allow for query and retrieval of symptoms, findings and disease features across the cohorts. Additional work is being done in collaboration with the INCLUDE (INvestigation of Co-occurring conditions across the Lifespan to Understand Down syndromE) project to augment the Kids First Data Resource Center’s ability to integrate survey data for assessment of Down Syndrome-related clinical data and neurobehavior.
Contact: Nicole Vasilevsky
Funding: NIH grant # 5U2CHL138346 and The Kids First supplement is funded by NIH grant # 3200670520-03S1
BRIDGE Center
The Integration, Dissemination and Evaluation (BRIDGE) Center for the Bridge to Artificial Intelligence (Bridge2AI) Program will engage the scientific community across the development lifecycle, from data to standards to tools and algorithms, in order to ensure that data are ethically sourced, machine understandable, and easily integrated with other data. This award supports the work of the Standards and Teaming Cores in the BRIDGE Center.
The NIH established the Bridge2AI, an ambitious program focused on the fairness of Artificial Intelligence applications. In this sense, “fairness” refers to AI that is accurate, explainable, generalizable to diverse patients, researchers, and other stakeholder groups. Bridge2AI aims to understand what lessons and approaches are reusable across AI-based projects and domains and thereby catalyze AI solutions to complex biomedical and behavioral health challenges. The BRIDGE Center project assembles multiple institutions into a nexus that will foster next-generation activities answering the myriad of socio-technological issues that surround biomedical/behavioral data collection and its usage with AI.
The BRIDGE Center Teaming Core will enable team science across Bridge2AI. The BRIDGE Center Standards Core aims to develop best practices to ensure that the standards implemented are as generalizable as possible in biomedical and behavioral applications (among other fields).
Together, the Teaming and Standards Core at the University of Colorado Anschutz Medical Campus will support innovation for the next generation of Artificial Intelligence and Machine Learning in relation to biomedicine, behavioral applications, and beyond.
Contact: Monica Munoz-Torres
Funding: NIH grant # U54 HG012513
Environmental Health Sciences Vocabulary
TISLab is laying the foundation for a new community-driven effort to build data standards for the environmental health sciences.
Contact: Anne Thessen
Funding: NIEHS
CD2H: Harmonizing the Informatics Community

TISLab is leading the National Center for Data to Health (CD2H) using team science and translational research to improve patient care. CD2H accelerates advancements in informatics by using findable, accessible, interoperable, and reusable (FAIR) principles to promote collaboration across the Clinical and Translational Science Awards (CTSA) Program community. CD2H tools and resources make it simple and valuable for CTSA Program members to get engaged, connect with peers, and contribute. By promoting collaboration, CD2H fosters a robust translational science informatics ecosystem that collectively develops solutions to solve clinical problems faster, more efficiently, and more effectively. CTSA Program members are poised to lead this charge by harnessing collective expertise and strengths to solve key informatics challenges.
Contact: Julie McMurry
Funding: NCATS grant # U24TR002306
Converging Genomics, Phenomics, and Environments Using Interpretable Machine Learning Models
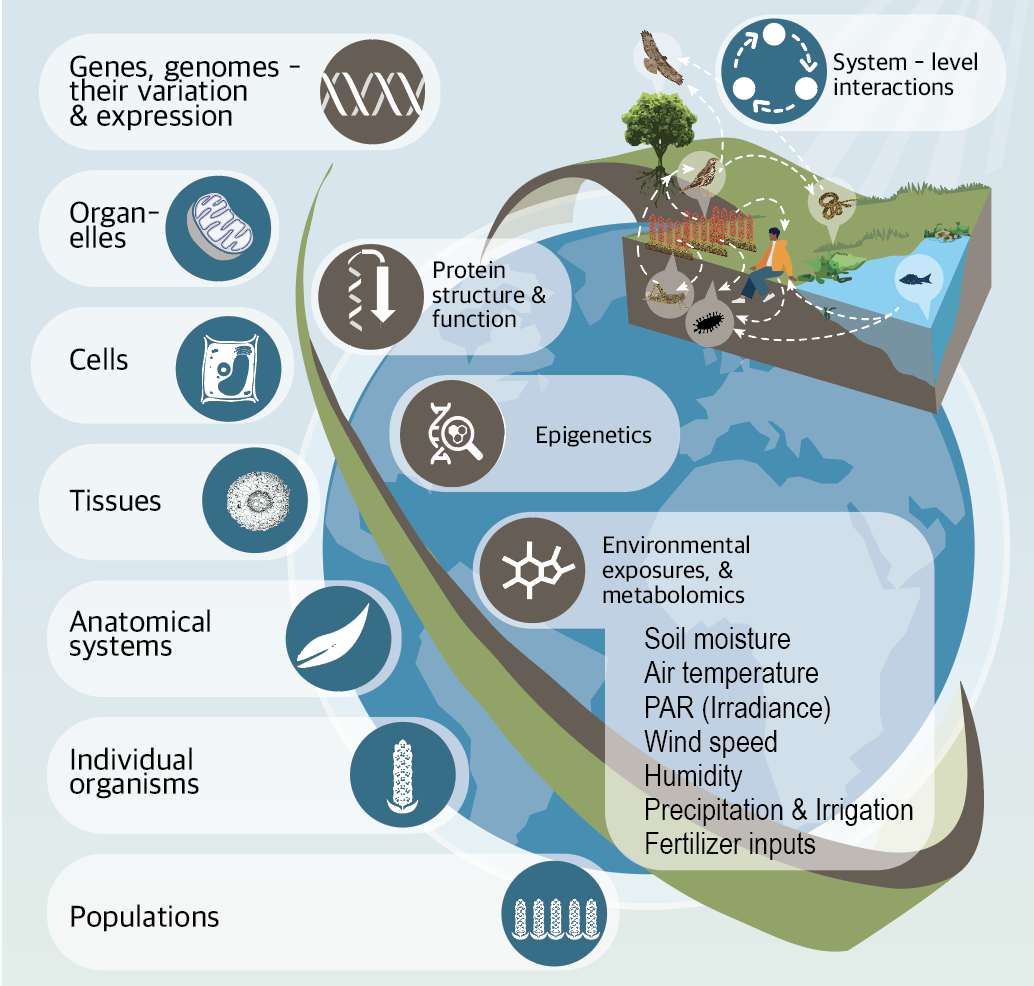
TISLab is revolutionizing our understanding of how genes are transformed into organism traits in a changing environment. One of the central dogmas of biology is that genes and environments work together to determine the characteristics of an organism, or its phenotype. Yet, being able to predict with precision what an organism will look like or do based on its genes and environment has, for the most part, remained out of reach. Turns out, there’s a lot going on in a cell and in an organism that we just don’t understand which has a huge impact on how that organism looks and behaves. In addition to revolutionizing the way we think about how genes are expressed, we have to develop brand new ways of transforming and analyzing data so we can make best use of innovations in artificial intelligence and deep learning. TISLab is leading this effort to innovate in computer science and biology to combat the negative effects of climate change on public health, conservation, and agriculture.
Contact: Anne Thessen
Funding: Office of Advanced Cyberinfrastructure (OAC), NSF Award #1940062
Integrated Health Sciences Facility Core

In collaboration with Oregon State’s Environmental Health Sciences Center the Linus Pauling Institute, TISLab is developing a new vision for the EHSC’s Integrated Health Sciences Facility Core, dedicated to connecting researchers and community stakeholders through technology and data. By building tools like an interactive, searchable Discovery Index to identify interest and expertise, we can quickly build cross-disciplinary teams to tackle new challenges, and connect existing teams to the communities their work impacts.
Contact: Anne Thessen
Funding: NIEHS 1P30ES030287-01A1
Forums for Integrative Phenomics
TISLab is bursting through communication barriers to advance science. One of the biggest barriers to scientific advancement is lack of communication across different kinds of expertise, especially between researchers who study genetics, environmental science, and phenomics. This results in data that are totally incompatible with each other and blocks scientific progress. The goal of this project is to bring together a diverse range of research scientists and clinicians from these three areas to find ways to communicate and share data. Unified data enables new insights in relationships between diseases, genes, and environments in a wide range of species. TISLab is creating new discoveries by planning and hosting workshops for creating new data standards that act as a “rosetta stone” between disciplines.
Contact: Julie McMurry
Funding: NIH grant # 1 U13 CA221044-01
Center for Cancer Data Harmonization

TISLab led the development of a data-powered support infrastructure to cure and treat cancer. The data that researchers need to develop new life saving therapies spans disciplines and biological scales, especially where personalized approaches are needed. Integrating data ranging from genomes and cellular components to patients and symptoms was a significant barrier to research.
To address these issues TISLab took a lead role in the Center for Cancer Data Harmonization (CCDH) whose mission is to bring data together across the various nodes of NCI’s Cancer Research Data Commons (CRDC) and make them accessible to researchers of any discipline in the fight against cancer.
Contact: Monica Munoz-Torres
Funding: NCI / Leidos contract # HHSN261201500003I
Undiagnosed Diseases Network (UDN)
We are part of the Metabolomics Core of the Undiagnosed Diseases Network (UDN). Our goals are to integrate metabolomics, lipidomics, glycomics, and genomics data with patient clinical phenotypes to provide mechanistic insight and aid diagnoses of rare and undiagnosed diseases. We are particularly involved in the integration of metabolites using existing pathway tools, reaction databases, and the integrated corpus of genotype-phenotype data within the Monarch platform for biological interpretation of disease etiology and biomarker signatures. We also started representing changes in glycomics signatures of patients with genetic diseases and undiagnosed diseases with the Molecular Glyco-Phenotype Ontology (MGPO) so as to enhance Human Phenotype Ontology and model data corpus to best leverage these phenotypic changes in the Exomiser tool.
Contact: Julie McMurry.
Phenotypr
Phenotypr is a free educational tool for people who believe they may have a disorder and want to learn more about their condition. This tool aims to provide additional information about what you are experiencing. We recommend you discuss this information with your healthcare provider to assist in your diagnosis.
Contact: Julie McMurry
Funding: PCORI grant # 1R24OD011883-01.
INCA Tools
This project aims to develop an intelligent concept assistant that will allow researchers to generate and share sets of metadata elements relevant to their project, and will use machine learning techniques to automatically apply this to data.
Contact: Melissa Haendel.
Funding: NHGRI grant # 5 U01 HG009453 02.
N-Lighten Network: A Federated Platform for Education Resource Sharing
Researchers at Harvard University, Oregon Health & Science University and The Ohio State University CTSA Program hubs are developing educational resources, tools and technologies and make them available online to trainees, investigators and other members of the translational scientific team.
Contact: Marijane White
National Cancer Institute Theasaurus (NCIt)
The NCI thesaurus (NCIt) is a widely used cancer reference taxonomy that covers over 100,000 terms, developed by the National Cancer Institute (NCI) as a standalone ontology since 2003. The NCI partnered with members of the Monarch Initiative to enhance the ontology for interoperability with OBO ontologies.
Contact: Nicole Vasilevsky
Funding: Leidos contract #15X143
OpenRIF
OpenRIF, the Open Research Information Framework, is an open source community devoted to representing expertise ecosystems - all the things we do and all the things we contribute. The community works on developing and promoting interoperable and extensible semantic infrastructure, such as the VIVO Integrated Semantic Framework (VIVO-ISF), an ontology for representing people, works, and the relationships between them; federated databases modeled on PARDI, the Portfolio Analysis and Reporting Data Infrastructure, for research impact and evaluation;and eagle-i, which aims to make research resources discoverable via a semantic search interface and represents their relationships to scholarly activities.
Contact: Robin Champieux
Annotating the CRAFT Corpus
The Colorado Richly Annotated Full-Text (CRAFT) Corpus is a collection of 97 full-length, open-access journal articles from the biomedical literature that are manually annotated, for use as gold-standard resources for the training and testing of biomedical Natural Language Processing (NLP) systems. Within these articles, each mention of the concepts explicitly represented in eight prominent Open Biomedical Ontologies (OBOs) has been annotated, resulting in gold-standard markup of genes and gene products, chemicals and molecular entities, biomacromolecular sequence features, cells and cellular and extracellular components and locations, organisms, biological processes and molecular functionalities. With these ~100,000 concept annotations among the ~800,000 words in the 67 articles of the 1.0 release, it is one of the largest gold-standard biomedical semantically annotated corpora. In addition to this substantial conceptual markup, the corpus is fully annotated along a number of syntactic and other axes, notably by sentence segmentation, tokenization, part-of-speech tagging, syntactic parsing, text formatting, and document sectioning. Current efforts are underway to add new annotations.
Contact: Nicole Vasilevsky
Funding: NIH grants 5R01LM008111 and 5R01LM009254, and DARPA-BAA-14-14.
Web Taxology project
The Web Taxology project is a collaboration between the OHSU Library, Digital Strategy, and the Marketing team to create a data model of all the people, places, and things at OHSU, which is the first step towards improving OHSU’s local search results in third-party search engines like Google. The initial goal of the project is to improve patient experience when finding their way to and around OHSU’s campuses and clinic locations, with future goals to be used in other contexts and projects throughout the institution as a whole.
Contact: Marijane White
Funding: OHSU Library
Open Insight
Open Insight is an education and outreach project designed to stimulate early career researchers’ engagement with open science practices through hands-on learning and conversations with leaders in the field. The Open Insight team brings together doctoral students, scientists, and OHSU Library staff with expertise in scholarly communications and data management to explore and promote the practice of open science activities and workflows.
Contact: Robin Champieux.
CTAR
The Clinical and Translational Activity Reporting (CTAR) tool was a collaboration Oregon Clinical and Translational Research Institute and the OHSU Library’s Ontology Development Group to prototype tool that would collocate and analyze data about research activities across a disparate set of internal and external databases (e.g. IRB, grants and contracts, PubMed). Leveraging MeSH, other terminologies, and simple Natural Language Processing (NLP) techniques, the CTAR prototype identified research activity topics and trends, and their classification as clinical or translational. The tool was intended to increase the OHSU’s and the Oregon Clinical and Translational Research Institute’s ability to strategically contribute to research outcomes and human health.
Contact: Melissa Haendel.
CTSAconnect
The CTSAconnect project aimed to integrate information about research activities, clinical activities, and scientific resources by creating an Integrated Semantic Framework (ontology). This new framework facilitated the production and consumption of Linked Open Data (a Semantic Web method of sharing data) about investigators, physicians, biomedical research resources, services, and clinical activities. The goal was to enable software to consume data from multiple sources and allow the broadest possible representation of researchers’ and clinicians’ activities and research products. Current research tracking and networking systems rely largely on publications, but clinical encounters, reagents, techniques, specimens, model organisms, etc., are equally valuable for representing expertise. CTSAconnect was a collaboration between members at OHSU, Stony Brook University, Cornell University, Harvard University, University at Buffalo, and the University of Florida, and leveraged the work of eagle-i, VIVO, and ShareCenter.
Contact: Nicole Vasilevsky
Funding: Booz Allen Hamilton grant # CTSA 10-001: 100928SB23
eagle-i Network
eagle-i is a free application that makes it easy to discover biomedical research resources at a growing network of universities; more than 50,000 resources are listed and more are added every week. Resource types include model organisms, reagents, core laboratory services, instrumentation, and biospecimens.
Contact: Julie McMurry
Funding: Booz Allen Hamilton grant # 90177520
Resource Identification Initiative
The Resource Identification Initiative (#RII) was designed to help researchers sufficiently cite the key resources used to produce the scientific findings reported in the biomedical literature. The project aimed to enable resource identification within the biomedical literature through a pilot study promoting the use of unique Research Resource Identifiers (RRIDs). In addition to being unique, RRID’s meet three key criteria, they are: 1) Machine readable; 2) Free to generate and access; 3) Consistent across publishers and journals. A diverse group of collaborators led the project, including the Neuroscience Information Framework and the OHSU Library.
Contact: Nicole Vasilevsky
Funding: NIH and the INCF
Biospecimen Query
This project explored options for enhancing search capabilities for an existing biospecimen search application. Text processing tools were used to map anatomy, pathology, and disease concepts from existing terminologies and ontologies to pathology reports that are currently represented in an unstructured natural text form. The concepts identified in the text were also organized in a relational structure to enable taxonomic and parthood based searches. This was a small exploratory project with a goal of integrating these capabilities in an ongoing effort to expand and integrate OHSU’s biospecimen databases.
Contact: Melissa Haendel
Funding: OHSU’s Medical Research Foundation
